Biomolecules
Maharashtra Board-Class-11-Science-Biology-Chapter-6
Notes-Part-2
|
Topics to be Learn : Part-1
Topics to be Learn : Part-2
|
Nucleic Acids :
Findings of Feulgen :
- In 1924, Feulgen showed that chromosomes contain DNA
- He found that nucleic acids contain two pyrimidine (cytosine and thymine) and two purine (adenine and guanine) bases.
Findings of Wilkins and coworkers :
The findings of Wilkins and coworkers were as follows:
- Purine and pyrimidine bases are placed regularly along the DNA molecules at a distance of 3.4 Å.
- DNA (Deoxyribonucleotide) is composed of sugar molecule (a pentose sugar of deoxyribose type), phosphoric acid (phosphates when in chemical combination), and nitrogen containing bases (nitrogen containing organic ring compounds).
- Bases are of two types: Pyrimidine bases and purine bases.
- Pyrimidine bases are single ring (monocyclic) nitrogenous bases. Cytosine, Thymine and uracil are pyrimidine.
- Purine are double ring (dicyclic) nitrogenous bases. Adenine and guanine are purines.
Implications proposed by Erwin Chargaff:
Chargaff analyzed the composition of DNA from various sources, and mention his implication from all his experiments.
- Purine and pyrimidine always occur in equal amount in DNA.
- The base ratio i.e. A+T/G+C may vary in the DNA of different groups of animals and plants but the ratio remains constant for particular species.
| Swiss biochemist, Friederich Miescher (1869) discovered and isolated nucleic acids from the pus cells. By 1938, it became evident that nucleic acids are of two types- deoxyribose nucleic acid (DNA) and ribose nucleic acid (RNA). DNA is found in chloroplasts and mitochondria. DNA is the hereditary material in most of the organisms. |
- The nucleic acids are among the largest of all molecules found in living beings. They contain three types of molecules a) 5 carbon sugar, b) Phosphoric acid and c) Nitrogen containing bases.
- Three join together to form a nucleotide of nucleic acid.
Structure of DNA :

DNA is a long chain made up of alternate sugar and phosphate groups. The sugar present in DNA is always a deoxyribose attached to a phosphate group. So, it forms a regular, repeating phosphate sugar sequence.
- A base is attached to sugar -phosphate chain. Together this unit which consist of sugar, phosphate and a base is called nucleotide.
- The nitrogenous base and a sugar of a nucleotide form a molecule called nucleoside. It lacks phosphate group. Four types of nucleoside are found in DNA molecule.
- In a nucleoside, nitrogenous base is attached to the first carbon atom (C-1) of the sugar and when a phosphate group gets attached with that of the carbon (C-5) atom of the sugar molecule a nucleotide molecule is formed.
- A single strand of DNA consists of several thousands of nucleotides one above the other.
- The phosphate group of the lower nucleotide attached with the 5th carbon atom of the deoxyribose sugar forms phospho-di-ester bond with that of the 3rd carbon atom of the deoxyribose sugar of the nucleotide placed just above it.
- Single long chain of polynucleotides of DNA consists of one end with sugar molecules not connected with another nucleotide having C-3 carbon which is not connected with phosphate group.
- The single polynucleotide strand of DNA is not straight but helical in shape.
- The DNA molecule consists of such two helical polynucleotide chains which are complementary to each other.
- The two complementary polynucleotide chains of DNA are held together by the weak hydrogen bonds.
- Adenine always pairs with thymine, and guanine with cytosine (a pyrimidine with a purine).
- Adenine-thymine pair consists of two hydrogen bonds and guanine-cytosine pair consists of three hydrogen bonds (Thus, if the sequence of bases of a polynucleotide chain is known, that of the other can be determined).
Structure of DNA molecule proposed by Watson and Crick :
Watson and Crick did not conduct any experiment on DNA. Crick was expert in physics, X-ray crystallography and Watson in viral and bacterial genetics. They only analyzed and comprehended the results of experiments performed by scientists like R. Franklin, M. Wilkins, etc.
- According to Watson and Crick, DNA molecule consists of two strands twisted around each other in the form of a double helix.
- The two strands i.e. polynucleotide chains are supposed to be in opposite direction so end of one chain having 3' lies beside the 5' end of the other.
- One turn of the double helix of the DNA measures about 34 Å.
- It consists paired nucleotides and the distance between two neighboring pair nucleotides is 3.4 Å
- The diameter of the DNA molecule has been found be 20 Å.
Ribonucleic Acid (RNA) : RNA stands for Ribonucleic Acid. It is a long single stranded polynucleotide chain which helps in protein synthesis, functions as a messenger and translates messages coded in DNA into protein.
- DNA found in the nucleus of a living cell has necessary message for the production of all enzymes required by it.
- DNA forms mRNA through the process of transcription. This mRNA through the process of translation forms proteins.
- Difference between DNA and RNA is because of sugar and base.
Structure of RNA :
- The other nucleic acid found in living organisms is Ribose nucleic acid.
- In most of the organisms it is not found to be hereditary material but in certain organisms like tobacco mosaic virus, it is the hereditary material.
- Like DNA, ribose nucleic acid also consists of polynucleotide chain with the difference that it consists of single ‘strand. Exceptions are Reovirus and wound tumor virus where RNA is double stranded.
- The nucleotides of RNA have ribose sugar instead of the deoxyribose sugar as in the case of DNA.
- In case of RNA; Uracil substitutes thymine of DNA.
- Purine, pyrimidine equality is not found in RNA molecule because of its single stranded structure.
- RNA strand is usually found folded upon itself in certain regions or entirely. These folding helps in stability of the RNA molecule.
- Most of the RNA polynucleotide chains start either with adenine or guanine.

Three types of cellular RNAs have been distinguished
- messenger RNA (mRNA) or template RNA,
- ribosomal RNA (rRNA),_
- transfer RNA (tRNA) or soluble RNA.
Messenger RNA (mRNA):
- It is a linear polynucleotide.
- It accounts 3% of cellular RNA.
- Its molecular weight is several million.
- mRNA molecule carrying information to form a complete polypeptide chain is called cistron
- Size of mRNA is related to the size of message it contains.
- Synthesis of mRNA begins at 5’ end of DNA strand and terminates at 3’ end.

Role of messenger RNA:
It carries genetic information from DNA to ribosomes, which are the sites of protein synthesis.
Ribosomal RNA (rRNA):
- rRNA was discovered by Kurland in 1960.
- It forms 50-60% part of ribosomes.
- It accounts 80-90% of the cellular RNA.
- It is synthesized in nucleus.
- It gets coiled at various places due to intrachain complementary base pairing.

Role of ribosomal RNA: It provides proper binding site for m-RNA during protein synthesis.
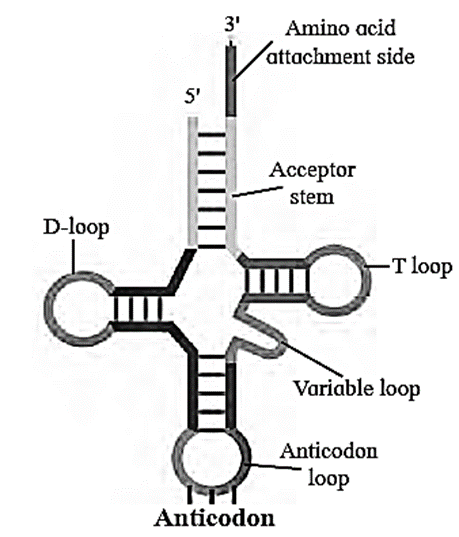
Transfer RNA (tRNA):
- These molecules are much smaller consisting of 70-80 nucleotides.
- Due to presence of complementary base pairing at various places, it is shaped like clover-leaf.
- Each tRNA can pick up particular amino acid.
- In the anticodon loop of tRNA, three unpaired nucleotides are present called as anticodon which pair with codon present on mRNA.
- The specific amino acids are attached at the 3' end in acceptor stem of clover leaf of tRNA.
Following four parts can be recognized on tRNA
- (i) DHU arm (Dihydroxyuracil loop/ amino acid recognition site)
- (ii) Amino acid binding site.
- (iii) Anticodon loop / codon recognition site
- (iv) Ribosome recognition site.
Role of transfer RNA: It helps in elongation of polypeptide chain during the process called translation.
| mRNA: The process of formation of m-RNA from DNA is called transcription.
rRNA: It is single stranded but folded upon itself in some regions. In some regions, nitrogen base pairing may be observed. tRNA: It is about 10 — 15% of the total cell RNA. |
Enzymes :
Enzymes are biological macromolecules which acts as a catalyst and accelerates the reaction in the body
- If enzymes were absent in the body, either the reactions would not occur or if they occur they would occure at a very slow rate.
- Each enzyme catalyzes only one reaction.
- Substrate : The substance upon which an enzyme acts is termed as the substrate.
- Endo-enzymes : The enzymes which act within the cell in which they are synthesized are known as endo-enzymes E.g enzymes produced in the chloroplast and mitochondria.
- Exo-enzymes : The enzymes which act outside the cell of which they are synthesized are known as exo-enzymes. E.g. enzymes released by many fungi. These enzymes, synthesized by living cell, retain their catalytic property even when extracted from cells.
On basis of chemical composition enzymes are categorised: _
- Purely proteinaceous enzymes: e.g. Proteases that spilt protein
- Conjugated enzymes: enzymes are made up of a protein to which a non-protein prosthetic group is attached.
Prosthetic group :
- Prosthetic group is non-protein in nature and is attached to the protein component of enzyme by chemical bonds.
- It is not removed by hydrolysis.
- If the prosthetic group is removed the protein part of the enzyme becomes inactive.
Coenzymes : Enzymes require certain organic compounds for their activity. The organic compounds that are tightly attached to the protein part are called coenzymes
- E.g. Nicotinamaide adenine dinucleotide (NAD), Flavin mononucleotide (FMN).
- Coenzymes always as organic molecule
Co-factors : Enzymes require certain inorganic ions for their activity.
The inorganic ions which are loosely attached to the protein part are called co-factors
- E. g. Magnesium, copper, zinc, iron, manganese etc.
Properties of enzyme :
Proteinaceous Nature:
- All enzymes are basically made up of protein.
Three-Dimensional conformation:
- All enzymes have specific 3-dimensional conformation.
- They have one or more active sites to which substrate (reactant) combines.
- The points of active site where the substrate joins with the enzyme is called substrate binding site.
Catalytic property:
- Enzymes are like inorganic catalysts and influence the speed of biochemical reactions but themselves remain unchanged.
- After completion of the reaction and release of the product they remain active to catalyze again.
- A small quantity of enzymes can catalyze the transformation of a very large quantity of the substrate into an end product.
- For example, sucrase can hydrolyze 100000 times of sucrose as compared with its own weight.
Specificity of action:
- The ability of an enzyme to catalyze one specific reaction and essentially no other is perhaps its most significant property. Each enzyme acts upon a specific substrate or a specific group of substrates.
Reversibility of action:
- Enzymes are very sensitive to temperature and pH.
- Each enzyme exhibits its highest activity at a specific pH i.e. optimum pH.
- Any increase or decrease in pH causes decline in enzyme activity e.g. enzyme pepsin (secreted in stomach) shows highest activity at an optimum pH of 2 (acidic).
- Trypsin (in duodenum) is most active at an optimum pH of 9.5 (alkaline).
- Both these enzymes viz. pepsin and trypsin are protein digesting enzymes.
Temperature:
- Enzymes are destroyed at higher temperature of 60-70°C or below, they are not destroyed but become inactive.
- This inactive state is temporary and the enzyme can become active at suitable temperature.
- Most of the enzymes work at an optimum temperature between 20°C and 35°C.
Nomenclature of Enzymes :
- Enzymes are named by adding the suffix- ‘ase’ to the name of the substrate on which they act e.g. protease, sucrase nuclease etc. which break up proteins, sucrose and nucleic acids respectively.
- The enzymes can be named according to the type of function they perform. For e.g., dehydrogenase remove hydrogen, carboxylase add CO; decarboxylases remove CO2, oxidases helping in oxidation.
- Some enzymes are named according to the source from which they are obtained. For e.g., papain from papaya, bromelain from the member of Bromeliaceae family, pineapple.
- According to international code of enzyme nomenclature, the name of each enzyme ends with an -ase an consists of double name.
- The first name indicates the nature of substrate upon which the enzyme acts and the second name indicates the reaction catalyzed. For e.g., pyruvic decarboxylase catalyses the removal of CO2 from the substrate pyruvic acid.
- Similarly, the enzyme glutamate pyruvate transaminase catalyses the transfer of an amino group from the substrate glutamate to another substrate pyruvate.
Classification of Enzymes :
Enzymes are classified into six classes:
(i) Oxidoreductases: These enzymes catalyze oxidation and reduction reaction by the transger of hydrogen and/or oxygen. e. g. alcohol dehydrogenase
Alcohol + NAD+ \(\frac {alocohole}{dehydogenase}\)> Aldehyde + NADH2
(ii) Transferases : These enzymes catalyse the transfer of certain groups between two molecules. e.g. glucokinase
Glucose + ATP \(\underrightarrow {Glucokinase}\) Glu-6-Phosphate + ADP
(iii) Hydrolases: These are enzymes catalyse hydrolytic reactions. This class includes amylases, proteases, lipases etc. eg. Sucrase
Sucrose \(\underrightarrow {Sucrase}\) Glucose + Fructose + H2O
(iv) Lyases : These enzymes are involved in elimination reactions resulting in the removal of a group of atoms from substrate molecule to leave a double bond.
It includes aldolases, decarboxylases, and dehydratases, e.g fumarate hydratase.
Histidine \(\frac {histidine}{decarboxylase}\)> Histamine + CO2
(v) Isomerases : These enzymes catalyze structural rearrangements within a molecule.
Their nomenclature is based on the type of isomerism. Thus these enzymes are identified as racemases, epimerases, isomerases, mutases, e.g. xylose isomerase.
Glu-6-Phosphate \(\underrightarrow {isomarase}\) Fructose-6-Phosphate
(vi) Ligases or Synthetases : These are the enzymes which catalyse the covalent linkage of the molecules utilizing the energy obtained from hydrolysis of an energy-rich compound like ATP, GTP e.g. glutathione synthetase, Pyruvate carboxylase.
Pyruvate + CO2 + ATP \(\frac {pyruvate}{carboxylase}\)> Oxaloacetate + ADP + Pi
Mechanism of enzyme action :
- The basic mechanism by which enzymes catalyze chemical reactions begins with the binding of the substrate (or substrates) to the active site on the enzyme.
- The active site is the specific region of the enzyme which combines with the substrate.
- The binding of the substrate to the enzyme causes changes in the distribution of electrons in the chemical bonds of the substrate and ultimately causes the reactions that lead to the formation of products.
- The products are released from the enzyme surface to regenerate the enzyme for another reaction cycle.
Models for enzyme actions : There are two types of models:
Lock and Key model:
- Lock and Key model was first postulated in 1894 by Emil Fischer.
- This model explains the specific action of an enzyme with a single substrate.
- In this model, lock is the enzyme and key is the substrate.
- The correctly sized key (substrate) fits into the key hole (active site) of the lock (enzyme).

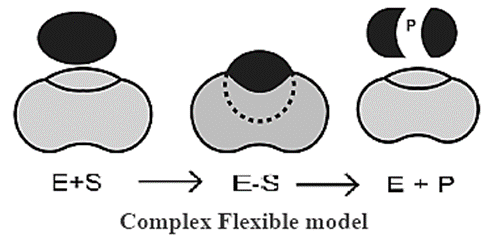
Induced Fit model (Flexible Model):
- Induced Fit model was first proposed in 1959 by Koshland.
- This model states that approach of a substrate induces a conformational change in the enzyme.
- It is the more accepted model to understand mode of action of enzyme. '
- The induced fit model shows that enzymes are rather flexible structures in which the active site continually reshapes by its interactions with the substrate until the time the substrate is completely bound to it.
- It is also the point at which the final form and shape of the enzyme is determined.
Factors affecting enzyme action :
The factors affecting the enzyme activity are as follows:
Concentration of substrate:
- Increase in the substrate concentration gradually increases the velocity of enzyme activity within the limited range of substrate levels.
- A rectangular hyperbola is obtained when velocity is plotted against the substrate concentration.
- Three distinct phases (A, B and C) of the reaction are observed in the graph.
- Where V = Measured velocity, Vmax = Maximum velocity, S = Substrate concentration, Km = Michaelis-Menten constant.
- Km or the Michaelis-Menten constant is defined as the substrate concentration (expressed in moles/lit) to produce half of maximum velocity in an enzyme catalyzed reaction.
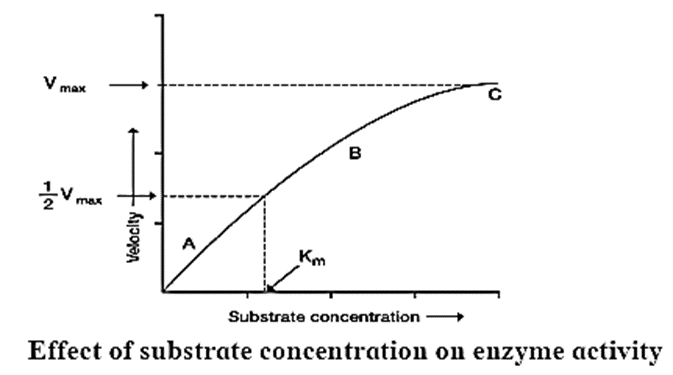
- It indicates that half of the enzyme molecules (i.e. 50%) are bound with the substrate molecules when the substrate concentration equals the Km value.
- Km value is a constant and a characteristic feature of a given enzyme.
- It is a representative for measuring the strength of ES complex.
- A low Km value indicates a strong affinity between enzyme and substrate, whereas a high Km value reflects a weak affinity between them.
- For majority of enzymes, the Km values are in the range of 10-5 to 10-2 moles.
Enzyme Concentration:
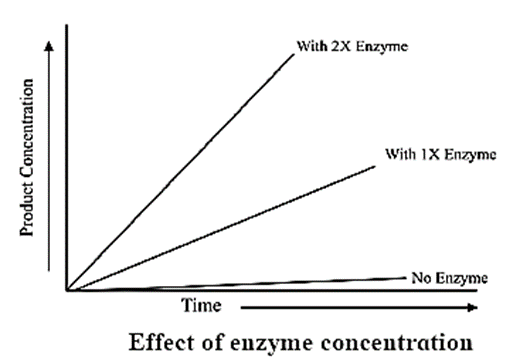
- The rate of an enzymatic reaction is directly proportional to the concentration of the substrate.
- The rate of reaction is also directly proportional to the square root of the concentration of enzymes.
- It means that the rate of reaction also increases with the increasing concentration of enzyme and the rate of reaction can also decrease by decreasing the concentration of enzyme.
Temperature:
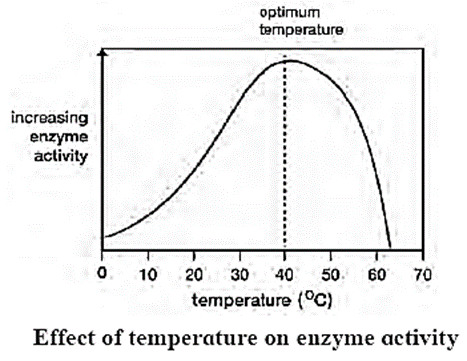
- The temperature at which the enzymes show maximum activity is called Optimum temperature.
- The rate of chemical reaction is increased by a rise in temperature but this is true only over a limited range of temperature.
- Enzymes rapidly denature at temperature above 40°C.
- The activity of enzymes is reduced at low temperature.
- The enzymatic reaction occurs best at or around 37°C which is the average normal body temperature in homeotherms(Organisms which can maintain their body temperature at constant level are known as homeotherms).
Effect of pH:

- The pH at which an enzyme catalyzes the reaction at the maximum rate is known as optimum pH.
- The enzyme cannot perform its' function beyond the range of its pH value.
Other substances:
- The enzyme action is also increased or decreased in the presence of some other substances such as coenzymes, activators and inhibitors.
- Most of the enzymes are combination of a co-enzyme and an apo-enzyme
- Activators are the inorganic substances which increase the enzyme activity.
- Inhibitor is the substance which reduces the enzyme activity.
Concept of Metabolism:
- Metabolism is the sum of the chemical reactions that take place within each cell of a living organism and provide energy for vital processes and for synthesizing new organic material.
- It involves continuous process of breakdown and synthesis of biomolecules through chemical reactions.
- Each of the metabolic reaction results in a transformation of biomolecules.
- Most of these metabolic reactions do not occur in isolation but are always linked with some other reactions.
In cells, metabolism involves two following types of pathways:
(i) Catabolic pathways :
This involves formation of simpler structure from a complex biomolecule.
For e.g. when we eat wheat, bread or chapati, our gastrointestinal tract digests (hydrolyses) the starch to glucose units with help of enzymes and releases energy in form of ATP (Adenosine triphosphate).
(ii) Anabolic pathway
It is also called as biosynthetic pathway that involves formation of a more complex biomolecules from a simpler structure.
For e.g., synthesis of glycogen from glucose and protein from amino acids. These pathways consume energy.

Metabolic pool :
- Metabolic pool is the reservoir of biomolecules in the cell on which enzymes can act to produce useful products as per the need of the cell.
- The concept of metabolic pool is significant in cell biology because it allows one type of molecule to change into another type E.g. Carbohydrates can be converted to fats and vice-versa.
- Metabolic pool in the cell is formed due to glycolysis and Krebs cycle.
- The catabolic chemical reaction of glycolysis and Krebs cycle provides ATP and biomolecules. These biomolecules form the metabolic pool of the cell.
- These biomolecules can be utilized for synthesis of many important cellular components.
- The metabolites can be added or withdrawn from the pool according to the need of the cell.
Metabolism : Metabolism is the sum of the chemical reactions that take place within each cell of a living organism and provide energy for vital processes and for synthesizing new organic material.
Secondary metabolites (SMs) :
- Biomolecules (organic compounds) like amino acids, proteins, lipids, sugars, etc. can also be called as metabolites.
- The above compounds are present in animal tissues and are called primary metabolites and their functions in normal biological processes are well known.
- The compounds other than primary metabolites found in plants, fungi and microbes are called as secondary metabolites.
Secondary metabolites are small organic molecules produced by organisms that are not essential for their growth, development and reproduction.
Secondary metabolites can be pigments (carotenoids, anthocyanins), alkaloids (morphine, codeine), terpenoides (Monoterpenes Diterpenes), toxins (Abrin, Ricin), drugs (vinblastin, curcumin), Lectins (concanavalin A), essential oils (lemon grass oil), polymeric substances (rubber, gums, cellulose), scents, Spices, antibiotics etc.
- Several types of bacteria, fungi and plants produce secondary metabolites.
- Secondary metabolites can be classified on the basis of chemical structure (e g SMs containing rings, sugar), composition (with or without nitrogen), their solubility in various solvents, or the pathway by which they are synthesized (e.g. phenylpropanoid produces tannins).
A simple way of classifying secondary metabolites includes three main groups such as:
- Terpenes: Made from mevalonic acid that is composed mainly of carbon and hydrogen
- Phenolics: Made from simple sugars containing benzene rings, hydrogen and oxygen.
- Nitrogen-containing compounds: Extremely diverse class may also contain sulphur.
Importance of secondary metabolites :
- Drugs developed from secondary metabolites have been used to treat infectious diseases, cancer, hypertension and inflammation.
- Morphine, the first alkaloid isolated from Papaver somniferum is used as pain reliver and cough suppressant.
- Secondary metabolites like alkaloids, nicotine, cocaine and the terpenes, cannabinol are widely used for recreation and stimulation.
- Flavours of secondary metabolites improve our food preferences.
- Tannins are added to Wines and chocolate for improving astringency.
- Since most secondary metabolites have antibiotic property, they are also used as food preservatives.
- Glucosinolates is a secondary metabolite which is naturally present in cabbage imparts a characteristic flavour and aroma because of nitrogen and sulphur containing chemicals. It also offers protection to these plants from many pests.
Main Page : – Maharashtra Board Class 11th-Biology – All chapters notes, solutions, videos, test, pdf.
Previous Chapter : Chapter-5-Cell Structure and Organization – Online Notes
Next Chapter : Chapter-7-Cell Division – Online Notes
We reply to valid query.