Biotechnology
Maharashtra Board-Class-12th-Biology-Chapter-12
Notes-Part-1
|
Topics to be Learn : Part-1
|
Biotechnology :
- The term biotechnology was first used by Karl Ereky in 1919 to describe a process for large scale production of pigs.
- Biotechnology is defined as ‘the development and utilization of biological forms, products or processes for obtaining maximum benefits to man and other forms of life’
- According to OECD (Organization for Economic Cooperation and Development, 1981)- ‘It is the application of scientific and engineering principles to the processing of materials by biological agents to provide goods and service to the human welfare’.
- Scientific and engineering principles : Scientific principles of microbiology, genetics biochemistry, chemical engineering, mathematics, statistics, computers, industrial processes, etc.
- Biological agent : Plants and animal cells, microorganisms, enzymes or their products.
Development of biotechnology in terms of its growth, occurred in two phases viz, Traditional biotechnology and Modern biotechnology.
Traditional or old biotechnology: Based on fermentation technology using microorganisms as in the preparation of curd, ghee, soma, vinegar, yogurt, cheese making, wine making, etc.
Modern or new biotechnology :
There are two major features of technology that differentiate modern biotechnology from classical or old biotechnology
These are,
- The use of r-DNA technology, polymerase chain reaction (PCR), microarrays, cell culture, cell fusion and bioprocessing to develop specific products.
- Ownership of technology and its socio political impact.
Principles and Processes of Biotechnology :
Modern biotechnology is based on two core techniques viz. genetic engineering and chemical engineering.
(i) Genetic engineering : Manipulation of genetic material towards a desired end and in a directed and predetermined way, using in vitro process. (ii) Chemical engineering: Maintaining sterile environment for manufacturing of useful products like vaccines, antibodies, enzymes, organic acids, vitamins, therapeutics, etc.
Technique of gene cloning and rDNA technology :
In gene cloning, a gene of known function can be transferred from its normal location into a cell (that of course does not contain it) via a suitable vector. The transferred gene is replicated normally and is handed over to the next progeny.
Tools and techniques for gene cloning/rDNA technology :
(i) The techniques used in r-DNA technology, on the basis of molecular weight : Gel permeation, osmotic pressure, ion exchange chromatography, spectroscopy, mass spectrometry, electrophoresis, etc.
(ii) Electrophoresis :
- It is used for the separation of charged molecules like DNA, RNA and proteins, by application of an electric field.
- Different types of electrophoresis : Agarose gel electrophoresis, PAGE, SDA PAGE.
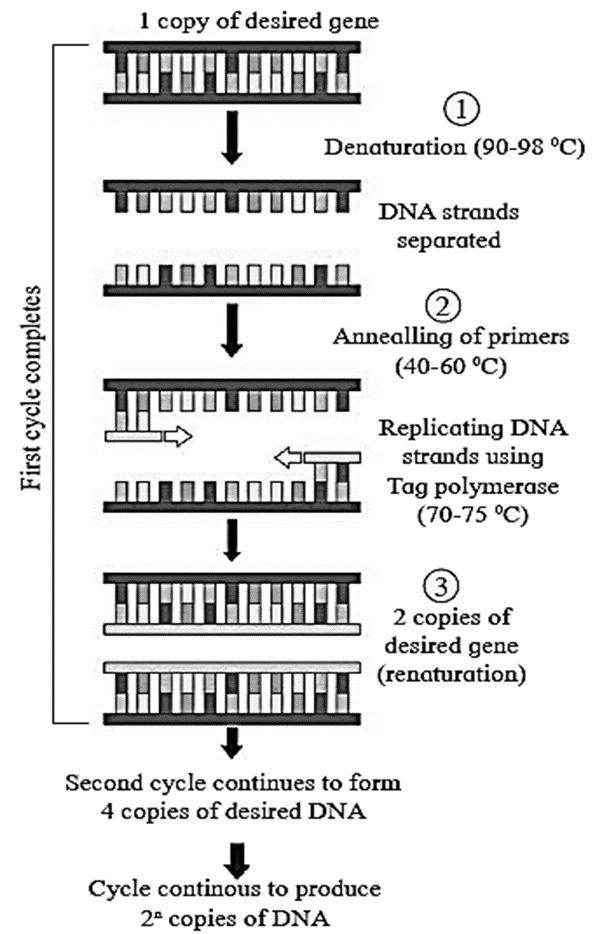
(iii) Polymerase chain reaction (PCR) :
It is used for in vitro gene cloning or gene multiplication to produce a billion copies of the desired segment of DNA or RNA, with high accuracy and specificity, in few hours.
The basic requirements of PCR technique are as follows :
- DNA containing the desired segment to be amplified.
- Excess of forward and reverse primers which are synthetic oligonucleotides of 17 to 30 nucleotide. They are complementary to the sequences present in DNA.
- dNTPs which are of four types such as dATP, dGTP, dTTP and dCTR
- A thermostable DNA polymerase (e.g. Taq DNA polymerase enzyme) that can withstand a high temperature of 90-98°C.
- Appropriate quantities of Mg++ ions.
- Thermal cycler, a device required to carry out PCR reactions.
Three essential steps in PCR : Denaturation, annealing of primer and extension of primer. One PCR cycle is of 3-4 minutes duration and it involves following steps :
Biological tools for gene cloning/r-DNA technology :
(1) Enzymes :
- Lysozymes, Nucleases (exonucleases, endonucleases, restriction endonucleases),
- DNA ligases, DNA polymerases, alkaline phosphatases, reverse transcriptases, etc.
- Nucleases : They cut the phosphodlester bonds of polynucleotide chains.
Types of nucleases :
- Exonucleases : They cut nucleotides from the ends of DNA strands.
- Endonucleases : They cut DNA from within.
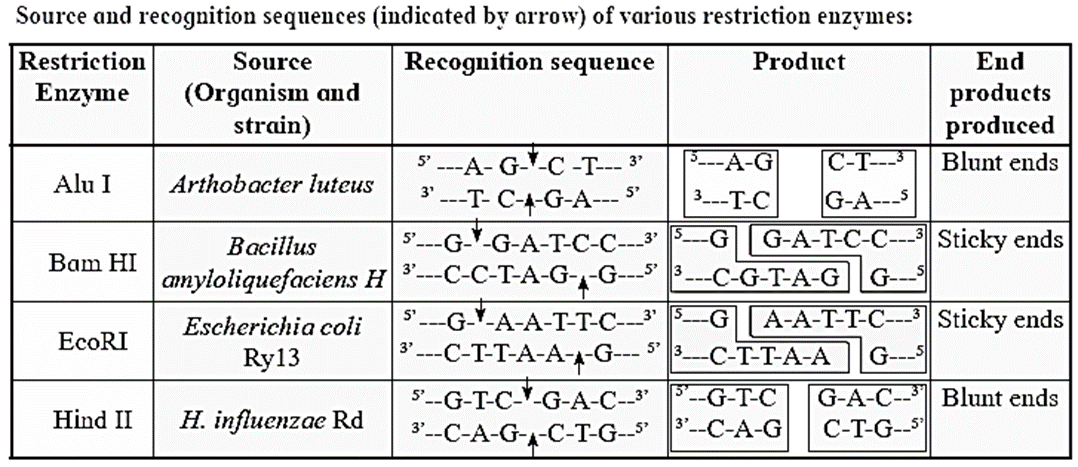
Restriction endonucleases or restriction enzymes :
- Restriction cutting may result in DNA fragments with blunt ends or cohesive or sticky ends or staggered ends (having short, single stranded projections).
- Restriction endonucleases have the ability to recognize specific sequences in DNA and cleave it.
- They are 4 to 8 nucleotides long and characterized by a particular type of internal symmetry.
- The specific site at which restriction endonuclease cuts the DNA is called recognition site or restriction site.
- Each restriction endonuclease recognizes its specific recognition sequence.
- For example, recognition sequence of by the enzyme EcoRI is
3’ ----C T T A A G -----5’
5' ----G A A T T C -----3’
- It is as palindrome. i.e. when read on opposite strand of DNA (3’ to 5’ or 5’ to 3’) it reads same.
- When the enzyme EcoRI recognizes this sequence, it breaks each strand at the same site in the sequence i.e. between the A and G residues.
- Restriction cutting may result in DNA fragments with blunt ends or cohesive or sticky ends or staggered ends [having short, single stranded projections).
Types of restriction enzyme :
- Type I : They function simultaneously as endonuclease and methylase e.g. EcoKI.
- Type II : They have separate cleaving and methylation activities e.g. EcoRI, BgII. They cut DNA at specific sites within the palindrome.
- Type III : They cut DNA at specific non-palindromic sequences e.g. HpaI, MboII.
(2) Cloning vectors (vehicle DNA) :
- Vectors are DNA molecules that carry a foreign DNA segment and replicate inside the host cell.
Examples of vectors : Plasmids (e.g. T1 plasmid of Agrobacterium tumefaciens, pBR 322, pUC), bacteriophages (e.g.M13, lambda virus), cosmid, phagemids, BAC (bacterial artificial chromosome), YAC (yeast artificial chromosome], transposons, baculoviruses and MACS (mammalian artificial chromosomes).
Plasmids as cloning vectors :

(3) Competent host :
- Cloning organism used are usually the bacteria like Bacillus haemophilus, Helicobacter pyroli and E.coli.
- Mostly E. coli is used for the transformation with recombinant DNA
Methodology for rDNA technology :
The steps involved in gene cloning are as follows : (i) Isolation of DNA (gene) from the donor organism : (ii) Insertion of desired foreign gene into a cloning vector (vehicle DNA) : (iii) Transfer of r-DNA into suitable competent host or cloning organism : (iv) Selection of the transformed host cell : (v) Multiplication of transformed host cell : (vi) Expression of gene to obtain desired product :

Notes, Solutions, Text Book-PDF
Class 12th-Biology-Chapter-12-Biotechnology-Text Book
Class 12th-Biology-Chapter-12-Biotechnology- Notes
Class 12th-Biology-Chapter-12-Biotechnology- Solution
PDF SET :
All Chapters Notes-Class-12-Biology-(15-PDF)-Maharashtra Board-Rs-130
All Chapters Solutions-Class-12-Biology-(15-PDF)-Maharashtra Board-Rs-128
All Chapters Notes+Solutions-Class-12-Biology-(30-PDF)-Maharashtra Board-Rs-240
Main Page : – Maharashtra Board Class 12th-Biology – All chapters notes, solutions, videos, test, pdf.
Previous Chapter : Chapter-11-Enhancement of Food production– Online Notes
Next Chapter : Chapter-13-Organisms and Population – Online Notes
We reply to valid query.